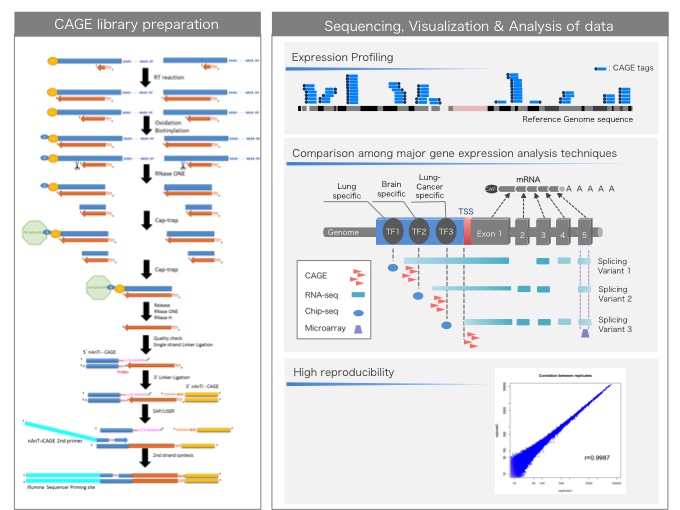
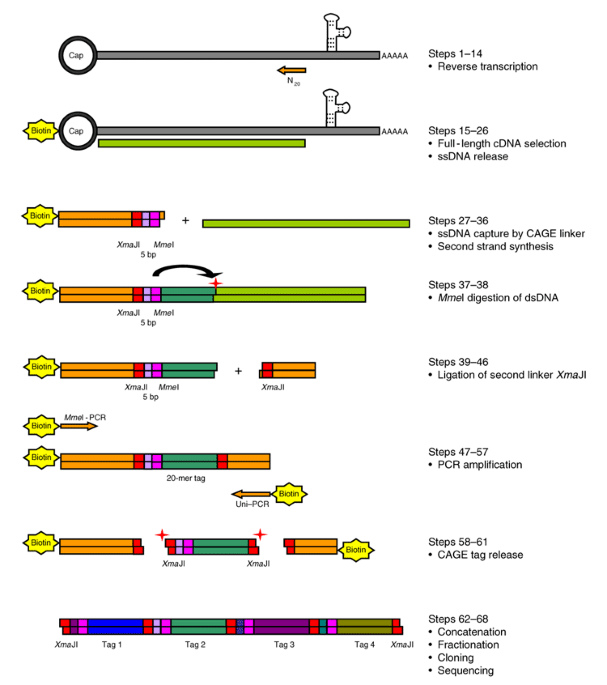
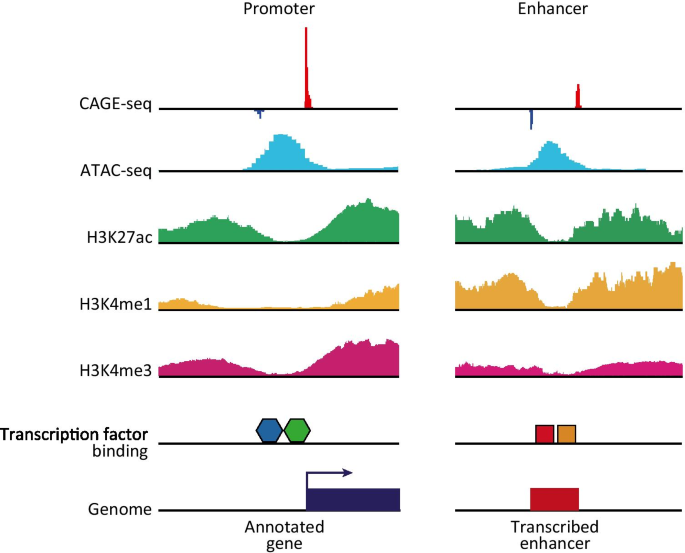
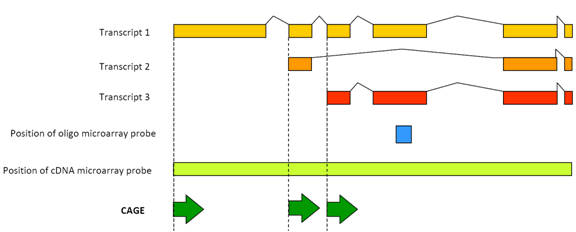
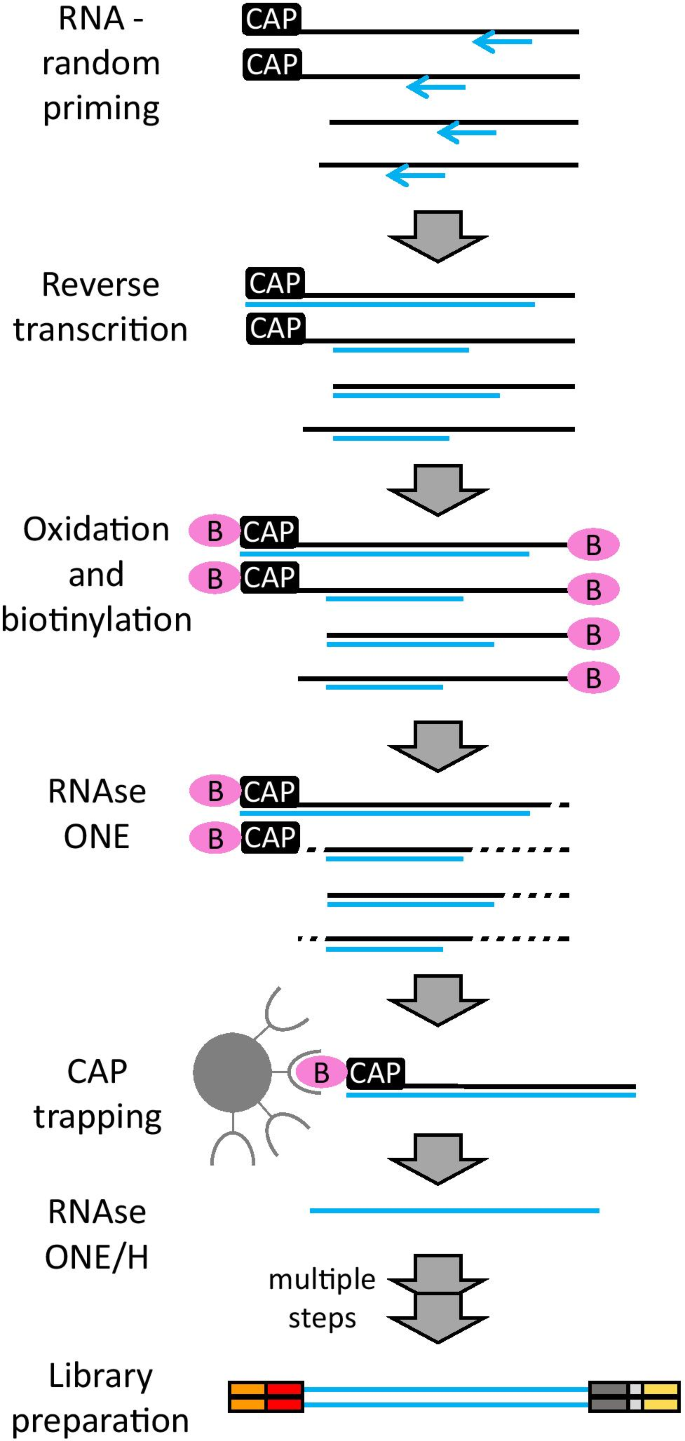
Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage | PNAS

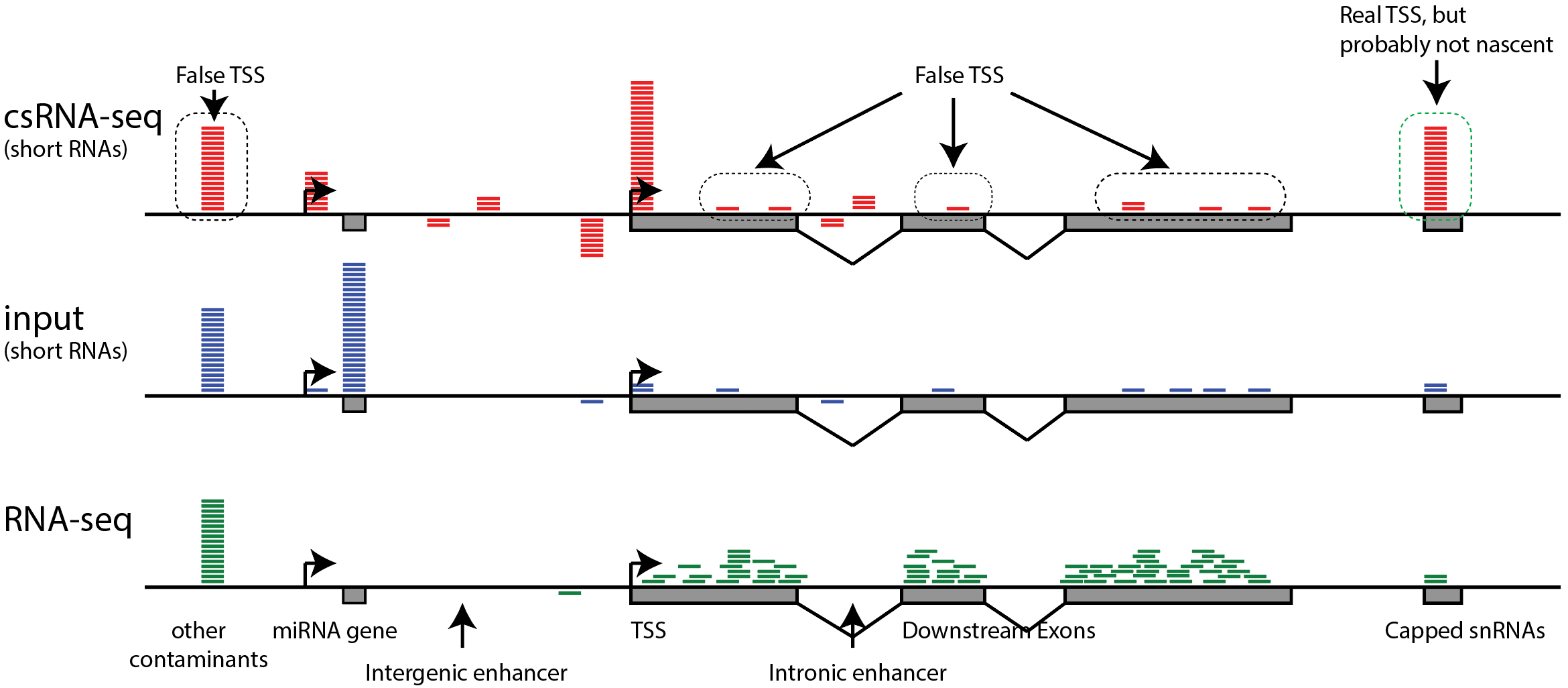
Characterization of Arabidopsis thaliana promoter bidirectionality and antisense RNAs by depletion of nuclear RNA decay enzymes | bioRxiv

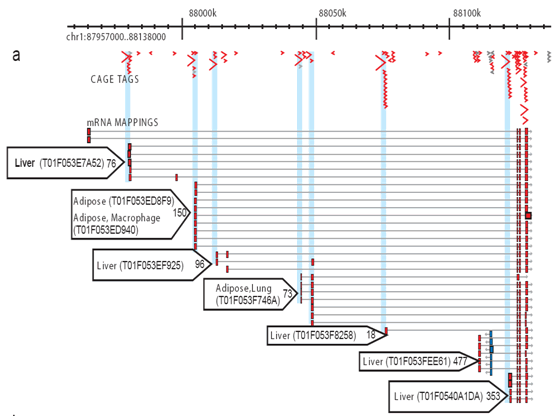
Cap analysis of gene expression reveals alternative promoter usage in a rat model of hypertension | Life Science Alliance

Solving the transcription start site identification problem with ADAPT-CAGE: a Machine Learning algorithm for the analysis of CAGE data | Scientific Reports
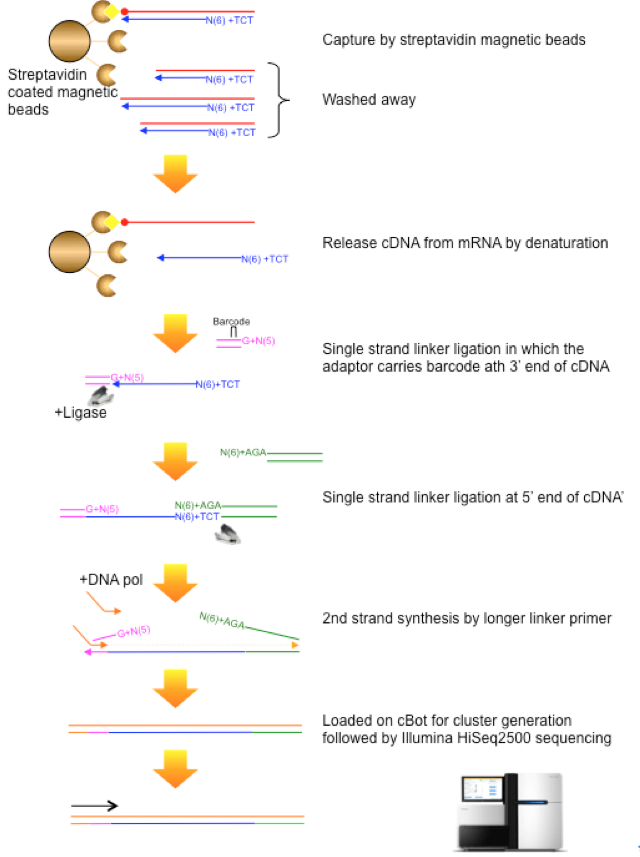
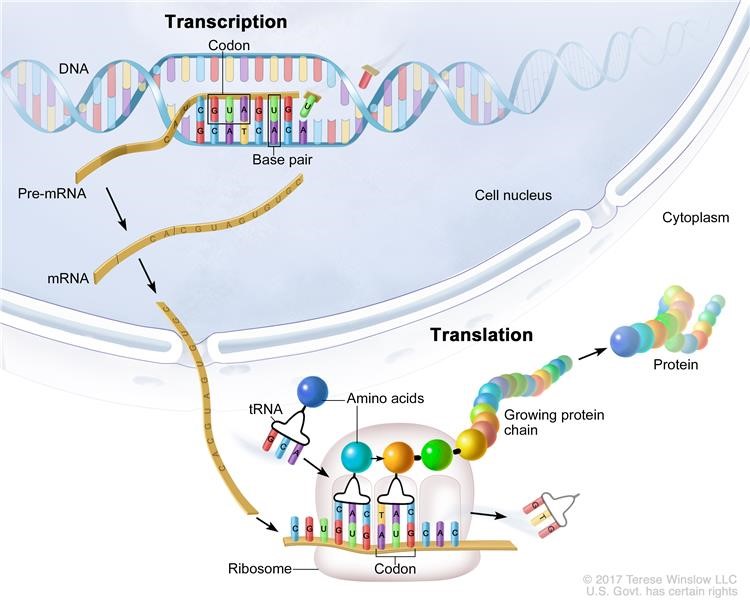
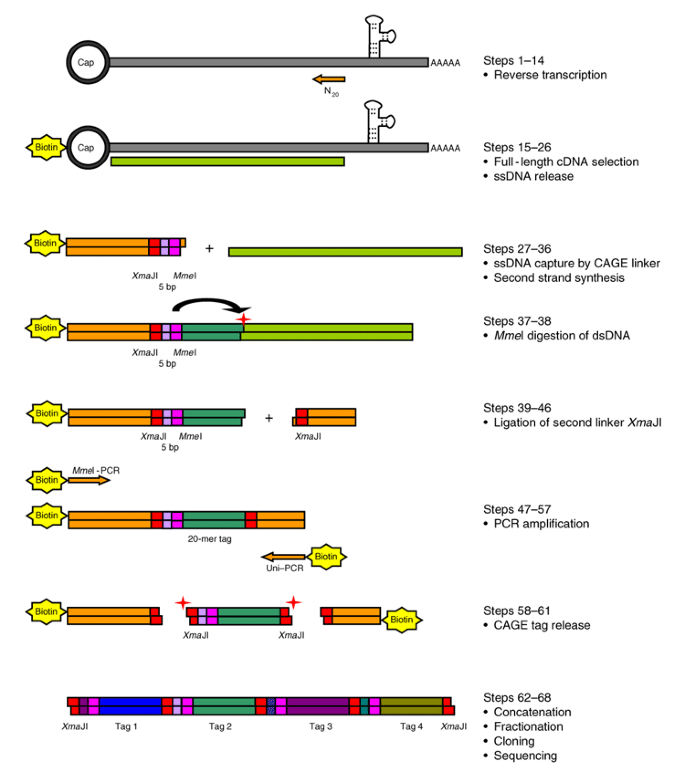
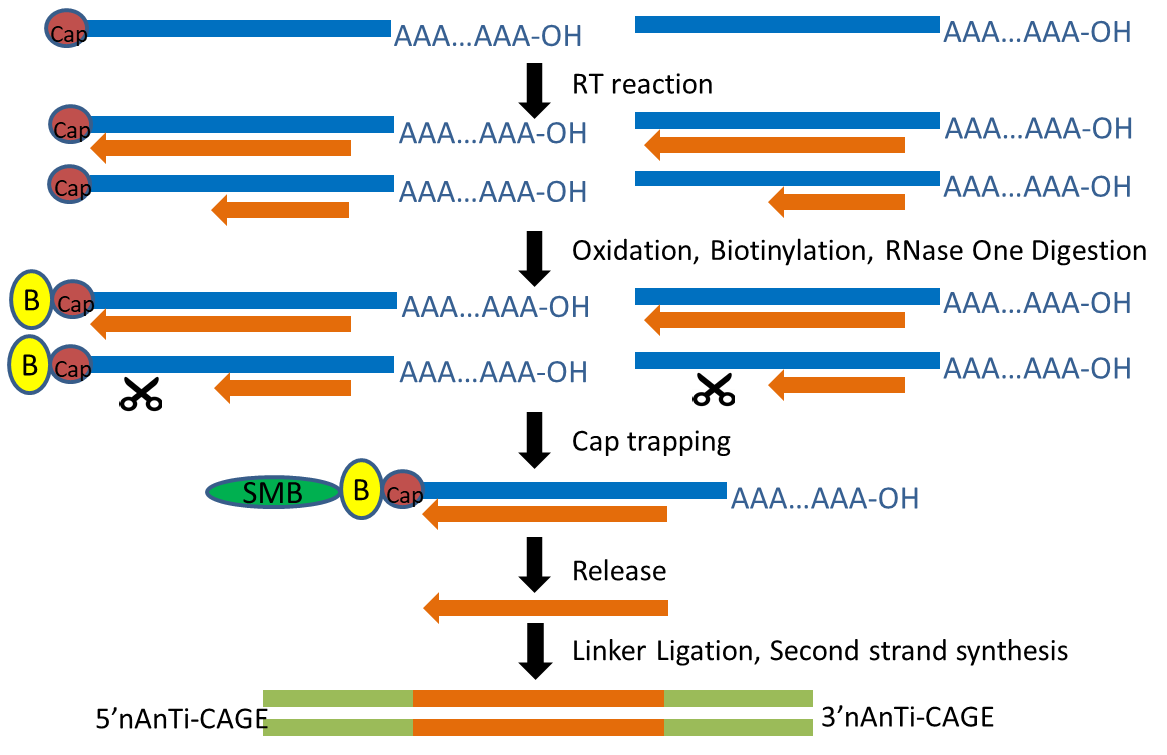
CAGE : A method for genome-wide identification of transcription start sites | 理化学研究所 ライフサイエンス技術基盤研究センター