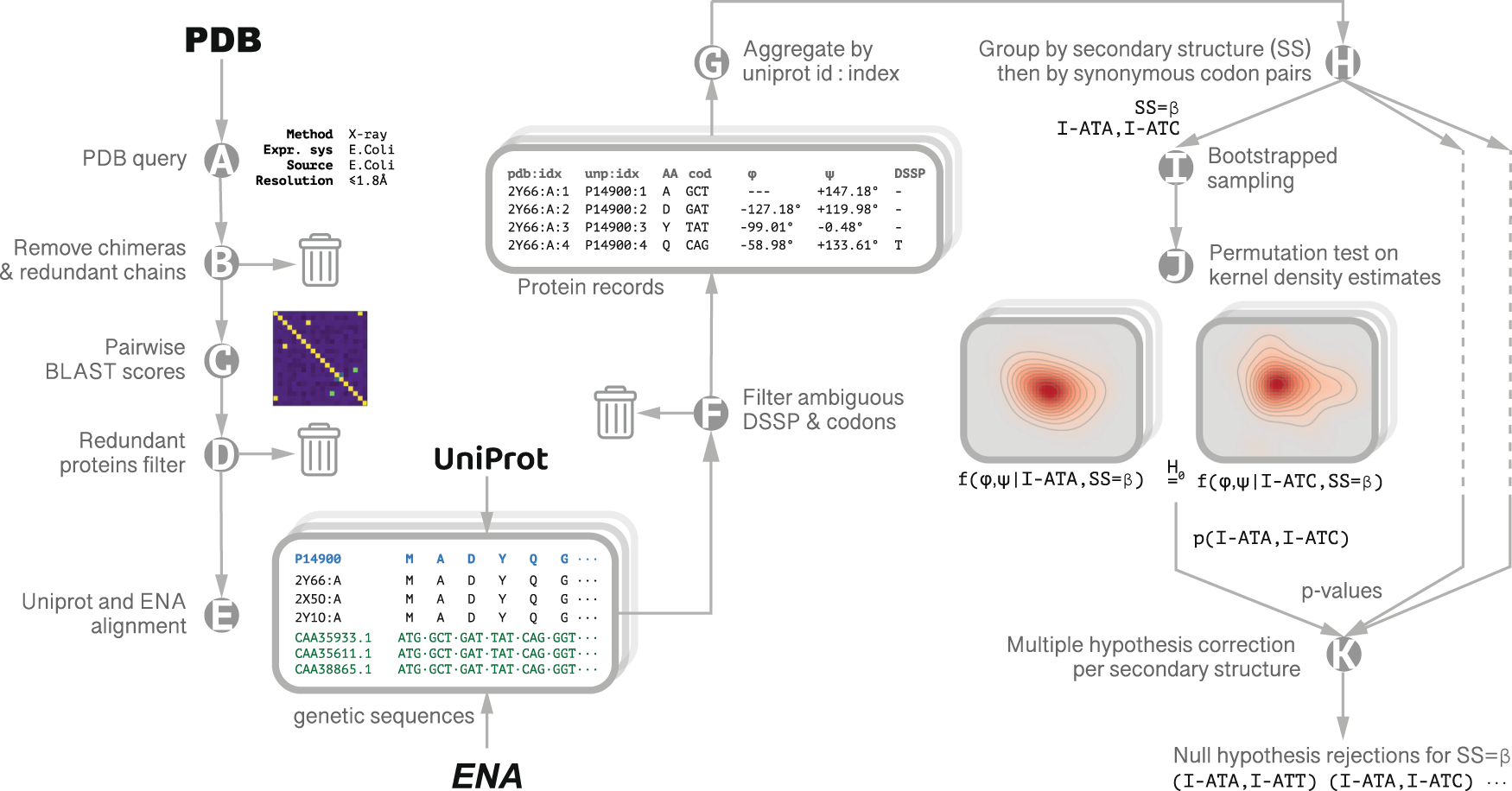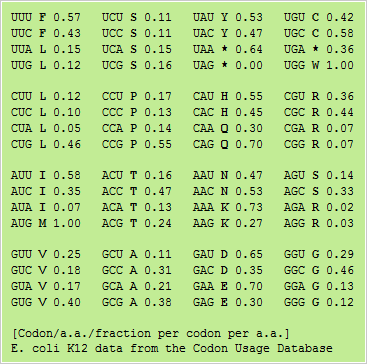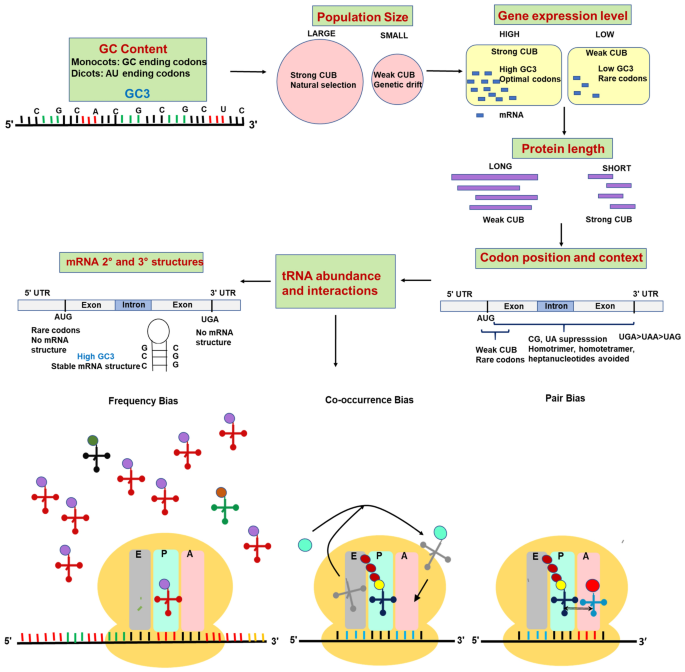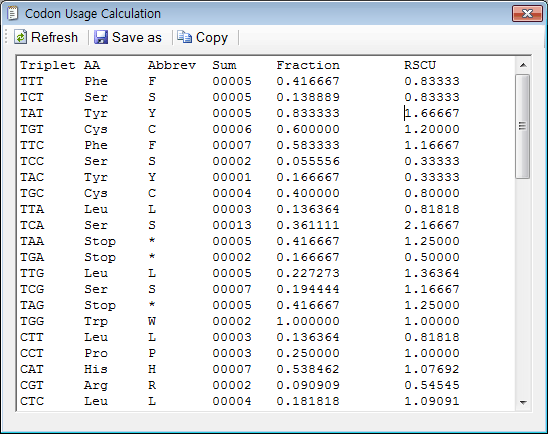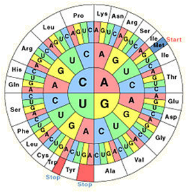
Frontiers | Codon Usage Is Influenced by Compositional Constraints in Genes Associated With Dementia
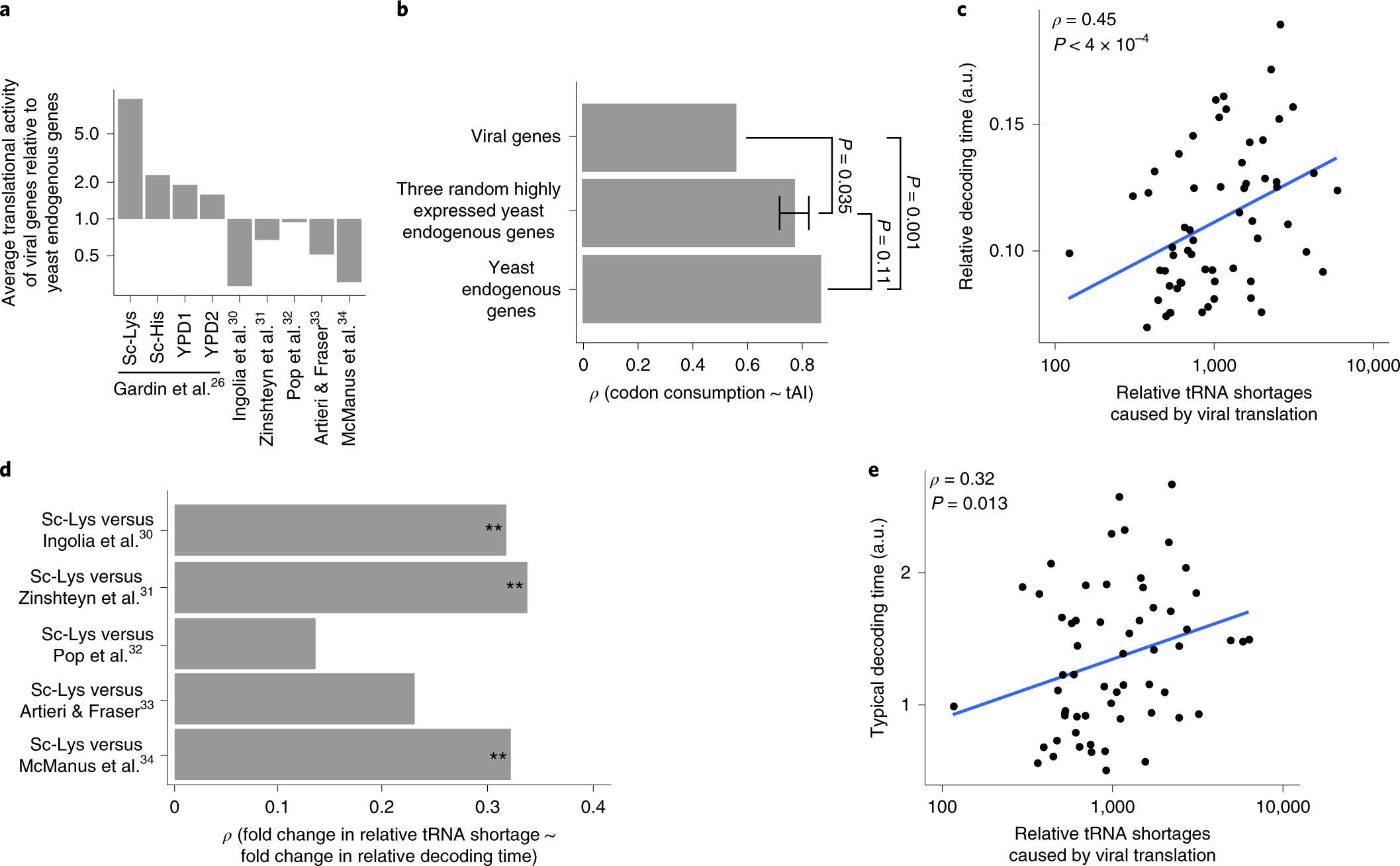
Dissimilation of synonymous codon usage bias in virus–host coevolution due to translational selection | Nature Ecology & Evolution
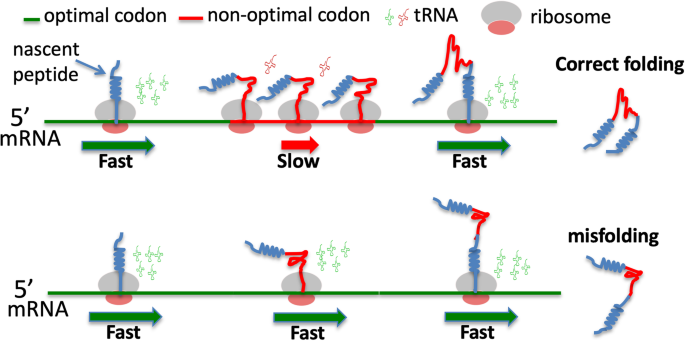
A code within the genetic code: codon usage regulates co-translational protein folding | Cell Communication and Signaling | Full Text
Calculating and comparing codon usage values in rare disease genes highlights codon clustering with disease-and tissue- specific hierarchy | PLOS ONE

MinMax: A versatile tool for calculating and comparing synonymous codon usage and its impact on protein folding - Rodriguez - 2018 - Protein Science - Wiley Online Library

Differences in codon bias and GC content contribute to the balanced expression of TLR7 and TLR9 | PNAS
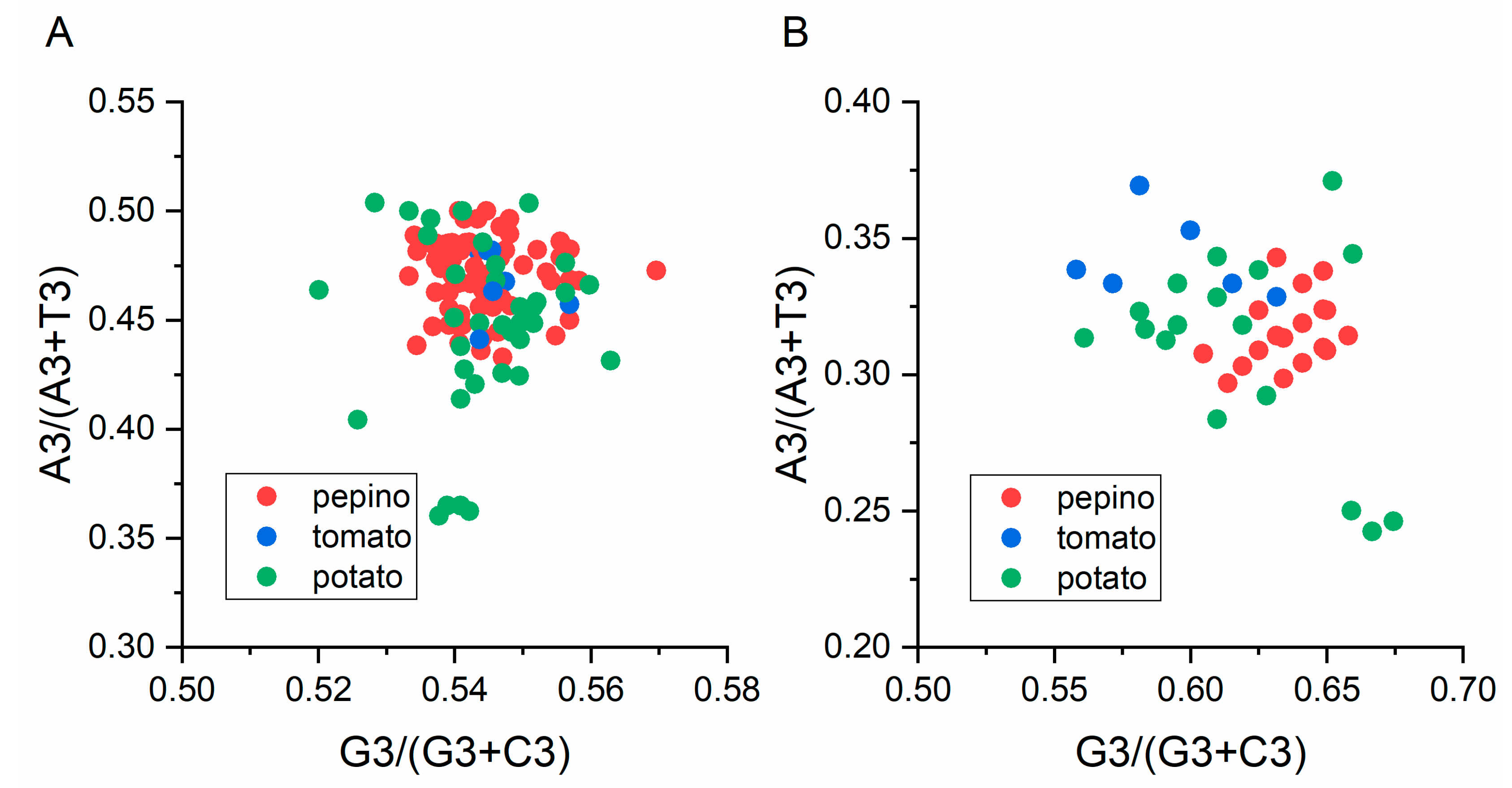
Viruses | Free Full-Text | Analysis of Synonymous Codon Usage Bias in Potato Virus M and Its Adaption to Hosts
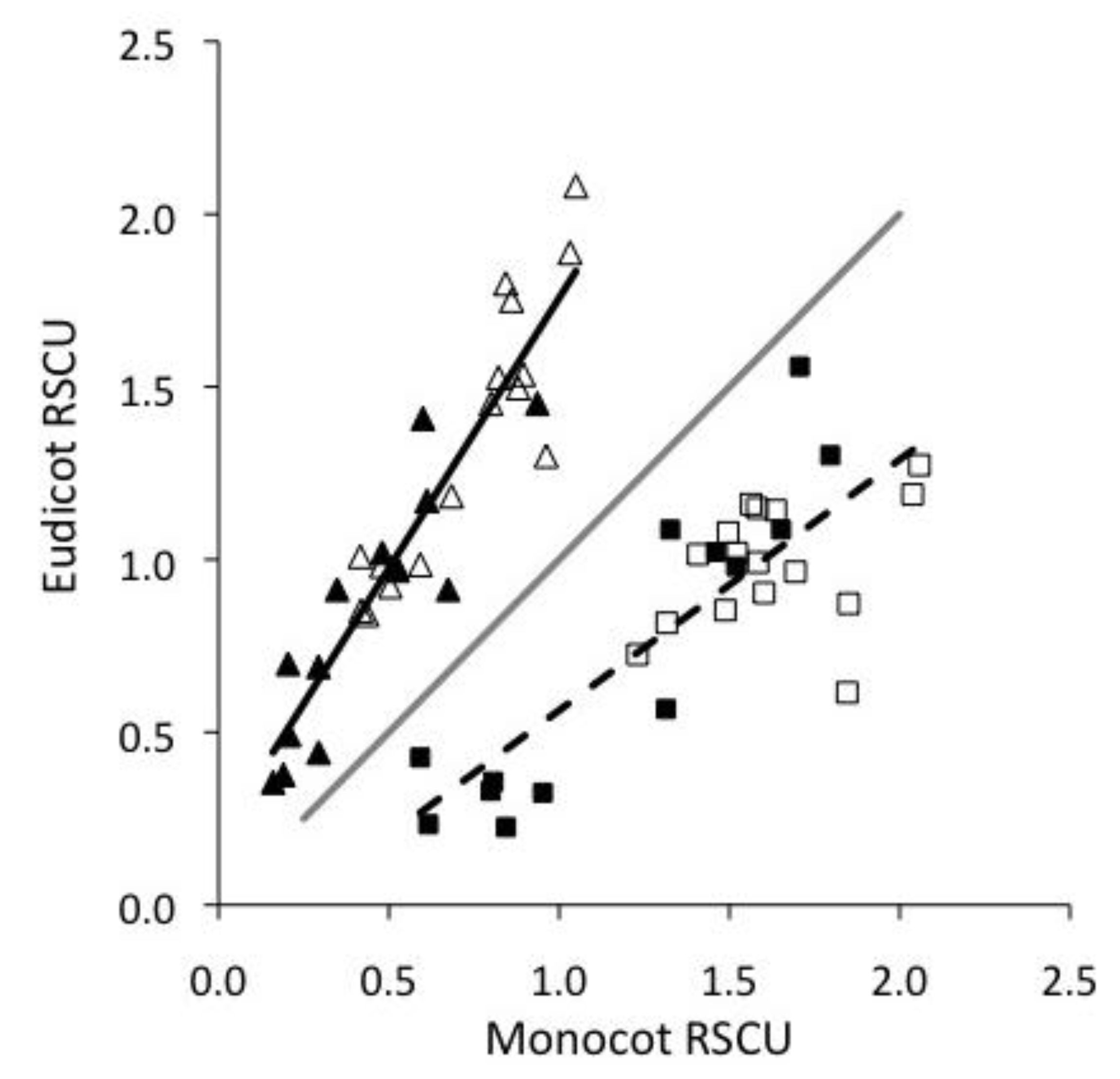
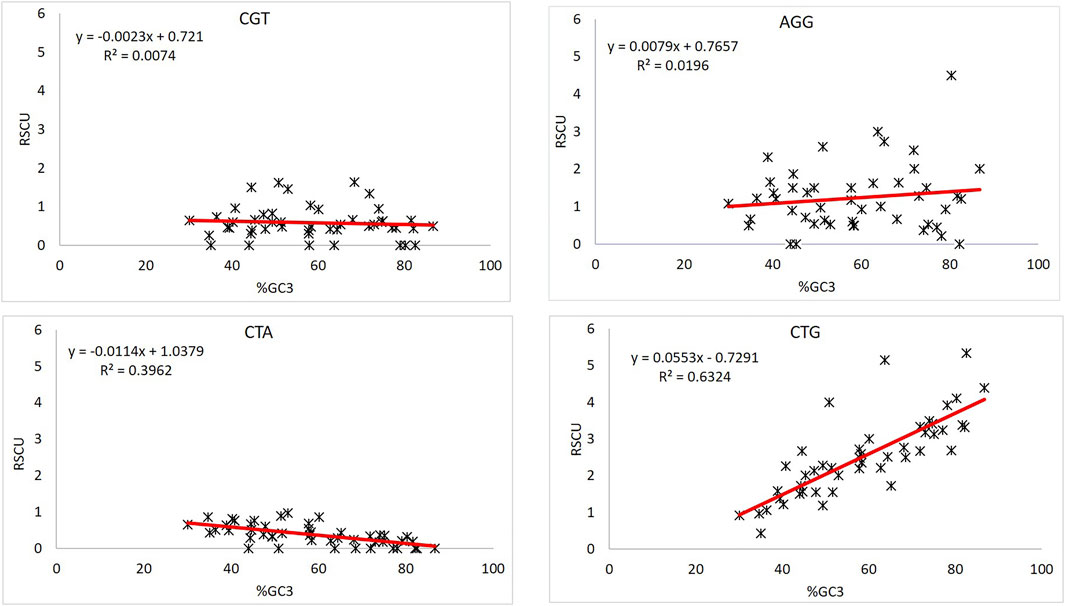
Viruses | Free Full-Text | Base Composition and Translational Selection are Insufficient to Explain Codon Usage Bias in Plant Viruses

Comparison of codon usage bias in model plant species using heat map of... | Download Scientific Diagram